
Chinese Journal OF Rice Science ›› 2017, Vol. 31 ›› Issue (4): 355-363.DOI: 10.16819/j.1001-7216.2017.6165 355
• Orginal Article • Previous Articles Next Articles
Liting SUN1,3, Tianzi LIN1,3, Yunlong WANG1, Mei NIU1, Tingting HU1, Shijia LIU1, Yihua WANG1, Jianmin WAN1,2,*( )
)
Received:2016-12-20
Revised:2017-02-07
Online:2017-07-25
Published:2017-07-10
Contact:
Jianmin WAN
孙立亭1,3, 林添资1,3, 王云龙1, 牛梅1, 胡婷婷1, 刘世家1, 王益华1, 万建民1,2,*( )
)
通讯作者:
万建民
基金资助:Liting SUN, Tianzi LIN, Yunlong WANG, Mei NIU, Tingting HU, Shijia LIU, Yihua WANG, Jianmin WAN. Phenotypic Analysis and Gene Mapping of a White Stripe Mutant st13 in Rice[J]. Chinese Journal OF Rice Science, 2017, 31(4): 355-363.
孙立亭, 林添资, 王云龙, 牛梅, 胡婷婷, 刘世家, 王益华, 万建民. 水稻白条纹突变体st13的表型分析及基因定位[J]. 中国水稻科学, 2017, 31(4): 355-363.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2017.6165 355
| 材料 Material | 株高 Plant height/cm | 有效穗数 No. of effective panicles | 剑叶长 Flag leaf length /cm | 穗长 Panicle length /cm | 结实率 Seed setting rate/% | 千粒重 1000-grain weight /g | |
|---|---|---|---|---|---|---|---|
| WT | 112.2±2.9 | 9.6±0.6 | 31.5±2.4 | 23.8±0.2 | 95.5±1.5 | 27.9±1.6 | |
| st13 | 70.8±4.7** | 5.8±0.5** | 31.9±1.5 | 17.9±0.5** | 74.0±1.8** | 20.3±0.9** | |
Table 1 Comparison of major agronomic traits between wild-type Dongjin and the st13 mutant.
| 材料 Material | 株高 Plant height/cm | 有效穗数 No. of effective panicles | 剑叶长 Flag leaf length /cm | 穗长 Panicle length /cm | 结实率 Seed setting rate/% | 千粒重 1000-grain weight /g | |
|---|---|---|---|---|---|---|---|
| WT | 112.2±2.9 | 9.6±0.6 | 31.5±2.4 | 23.8±0.2 | 95.5±1.5 | 27.9±1.6 | |
| st13 | 70.8±4.7** | 5.8±0.5** | 31.9±1.5 | 17.9±0.5** | 74.0±1.8** | 20.3±0.9** | |
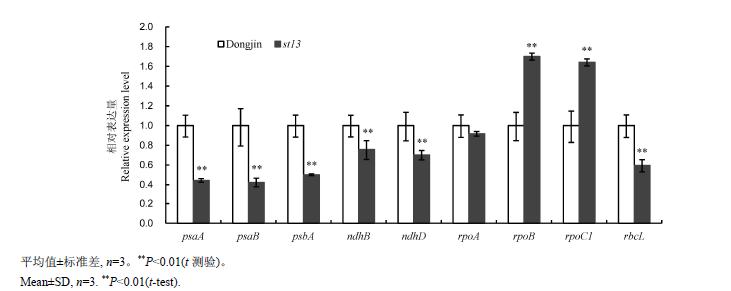
Fig. 4. Quantitative RT-PCR analyses of genes associated with chloroplast development and photosynthetic system in wild-type Dongjin and st13 mutant at the seedling stage.
| 组合 Combination | 正常株数 No. of normal plants | 白条纹株数 No. of plants with white stripes | 实际分离比 Segration ratio | χ2(3:1) | χ20.05 |
|---|---|---|---|---|---|
| Dongjin×st13 | 286 | 98 | 2.92:1 | 0.03 | 3.84 |
| st13×Dongjin | 281 | 89 | 3.16:1 | 0.13 |
Table 2 Segregation of F2 populations from wild-type Dongjin and st13 mutant.
| 组合 Combination | 正常株数 No. of normal plants | 白条纹株数 No. of plants with white stripes | 实际分离比 Segration ratio | χ2(3:1) | χ20.05 |
|---|---|---|---|---|---|
| Dongjin×st13 | 286 | 98 | 2.92:1 | 0.03 | 3.84 |
| st13×Dongjin | 281 | 89 | 3.16:1 | 0.13 |
| 引物名称 Primer name | 前引物 Forward primer | 后引物 Reverse primer | BAC克隆 BAC clone | |
|---|---|---|---|---|
| I3-21 | GCGAGATGGGCAGCTACTAC | ACACAATGTCCAGCTTGCAG | OSJNBa0010D22 | |
| P5 | CCTCCTCATAGCCCCAATCC | TCACTCTCCCATAGGTGGTC | OSJNBb0042N11 | |
| P3-13 | CCTCCGCACGAACCCT | GTGGGAAACTGGAGACGAG | OSJNBa0087M10 | |
| P3-14 | GGTTACATCTCCTTTTCGTTT | TCTTTGTTTGCTGCCATCT | OJ1519_A12 | |
| S-57 | AAAACCAAAACTAGAAGC | ATGTCCCTGGAAATGTA | OSJNBa0004G03 | |
| S-24 | CTCCGAGCGGCTGATTA | GGATAGTACTTCGTTTCGTA | OSJNBa0004G03 | |
| P3-29 | CAAACGCAATCTCACATCACA | AAGCGAGTGGGAGGGTG | OSJNBa0091E13 | |
| I3-22 | AGGTCTCGTGTCGTTCATCC | TGGAGGGAGCATGTCTATCA | OSJNBa0063J18 | |
Table 3 Polymorphic InDel markers used in this study.
| 引物名称 Primer name | 前引物 Forward primer | 后引物 Reverse primer | BAC克隆 BAC clone | |
|---|---|---|---|---|
| I3-21 | GCGAGATGGGCAGCTACTAC | ACACAATGTCCAGCTTGCAG | OSJNBa0010D22 | |
| P5 | CCTCCTCATAGCCCCAATCC | TCACTCTCCCATAGGTGGTC | OSJNBb0042N11 | |
| P3-13 | CCTCCGCACGAACCCT | GTGGGAAACTGGAGACGAG | OSJNBa0087M10 | |
| P3-14 | GGTTACATCTCCTTTTCGTTT | TCTTTGTTTGCTGCCATCT | OJ1519_A12 | |
| S-57 | AAAACCAAAACTAGAAGC | ATGTCCCTGGAAATGTA | OSJNBa0004G03 | |
| S-24 | CTCCGAGCGGCTGATTA | GGATAGTACTTCGTTTCGTA | OSJNBa0004G03 | |
| P3-29 | CAAACGCAATCTCACATCACA | AAGCGAGTGGGAGGGTG | OSJNBa0091E13 | |
| I3-22 | AGGTCTCGTGTCGTTCATCC | TGGAGGGAGCATGTCTATCA | OSJNBa0063J18 | |
| 基因 | 功能注释 |
|---|---|
| Gene | Annotation |
| Os03g0603300 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0603500 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0603600 | 甘油磷酸二酯酶 Glycerophosphoryl diester phosphodiesterase family protein |
| Os03g0603700 | A22A肽酶 Peptidase A22A, presenilin family protein |
| Os03g0604200 | 尿苷二磷酸葡萄糖脱氢酶 Similar to UDP-glucose dehydrogenase |
| Os03g0604500 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0604600 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0604700 | 假定蛋白 Hypothetical protein |
Table 4 Predicted genes and their putative functions in the 94 kb region.
| 基因 | 功能注释 |
|---|---|
| Gene | Annotation |
| Os03g0603300 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0603500 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0603600 | 甘油磷酸二酯酶 Glycerophosphoryl diester phosphodiesterase family protein |
| Os03g0603700 | A22A肽酶 Peptidase A22A, presenilin family protein |
| Os03g0604200 | 尿苷二磷酸葡萄糖脱氢酶 Similar to UDP-glucose dehydrogenase |
| Os03g0604500 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0604600 | 保守假定蛋白 Conserved hypothetical protein |
| Os03g0604700 | 假定蛋白 Hypothetical protein |
| [1] | Fambrini M, Castagna A, Dalla V F, Degl’Innocenti E, Ranieri A, Vernieri P, Pardossi A, Guidi L, Rascio N, Pugliesi C. Characterization of a pigment-deficient mutant of sunflower (Helianthus annuus L.) with abnormal chloroplast biogenesis, reduced PSII activity and low endogenous level of abscisic acid. Plant Breeding, 2004, 6: 645-650. |
| [2] | Agrawal G K, Yamazaki M, Kobayashi M, Hirochika R, Miyao A, Hirochika H.Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion: Tagging of a zeaxanthin epoxidase gene and a novel ostatc gene. Plant Physiol, 2001, 125(3): 1248-1257. |
| [3] | Parks B M, Quail P H.Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell, 1991, 3(11): 1177-1186. |
| [4] | Kusumi K, Sakata C, Nakamura T, Kawasaki S, Yoshimura A, Iba K.A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions.Plant J, 2011, 68(6): 1039-1050. |
| [5] | Lee S, Kim J H, Yoo E S, Lee C H, Hirochika H, An G.Differential regulation of chlorophyll a oxygenase genes in rice.Plant Mol Biol, 2005 57: 805-818. |
| [6] | Sugimoto H, Kusumi K, Noguchi K, Yano M, Yoshimura A, Iba K.The rice nuclear gene,VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J, 2007, 52: 512-527. |
| [7] | Zhang H, Li J, Yoo J, Yoo S, Cho S, Koh H, Seo H S, Paek N.Rice chlorina-1 and chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol, 2006, 62: 325-337. |
| [8] | Jung K H, Hur J, Ryu C H, Choi Y, Chung Y Y, Miyao A, Hirochika H, An G.Characterization of a rice chlorophyll- deficient mutant using the T-DNA gene-trap system.Plant Cell Physiol, 2003, 44: 463-472. |
| [9] | Kong W, Yu X, Chen H, Liu L, Xiao Y, Wang Y, Wang C, Lin Y, Yu Y, Wang C, Jiang L, Zhai H, Zhao Z, Wan J.The catalytic subunit of magnesium-protoporphyrin IX monomethyl ester cyclase forms a chloroplast complex to regulate chlorophyll biosynthesis in rice.Plant Mol Biol, 2016, 92(1-2): 177-191. |
| [10] | Wang P, Gao J, Wan C, Zhang F, Xu Z, Huang X, Sun X, Deng X.Divinyl chlorophyll (ide) a can be converted to monovinyl chlorophyll (ide) a by a divinyl reductase in rice.Plant Physiol, 2010, 153: 994-1003. |
| [11] | Sakuraba Y, Rahman M L, Cho S H, Kim Y S, Koh H J, Yoo S C, Paek N C.The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J, 2013, 74: 122-133. |
| [12] | Wu Z M, Zhang X, He B, Diao L, Sheng S, Wang J, Guo X, Su N, Wang L, Jiang L, Wang C, Zhai H, Wan J.A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis.Plant Physiol, 2007, 145: 29-40. |
| [13] | Yang Y, Xu J, Huang L, Leng Y, Dai L, Rao Y, Chen L, Wang Y, Tu Z, Hu J, Ren D, Zhang G, Zhu L, Guo L, Qian Q, Zeng D.PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J Exp Bot, 2016, 67(5): 1297-1310. |
| [14] | Sato Y, Morita R, Katsuma S, Nishimura M, Tanaka A, Kusaba M.Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice.Plant J, 2009, 57: 120-131. |
| [15] | Wang L, Wang C, Wang Y, Niu M, Ren Y, Zhou K, Zhang H, Lin Q, Wu F, Cheng Z, Wang J, Zhang X, Guo X, Jiang L, Lei C, Wang J, Zhu S, Zhao Z, Wan J.WSL3, a component of the plastid-encoded plastid RNA polymerase, is essential for early chloroplast development in rice.Plant Mol Biol, 2016, 92(4-5): 581-595. |
| [16] | Wang Y, Wang C, Zheng M, Lyu J, Xu Y, Li X, Niu M, Long W, Wang D, Wang H Y, William T, Wang Y, Wan J.WHITE PANICLE1, a Val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development.Plant Physiol, 2016, 170(4): 2110-2123. |
| [17] | Lopez-Juez E.Plastid biogenesis between light and shadows.J Exp Bot, 2007, 58: 11-26. |
| [18] | Sakamoto W, Miyagishima S Y, Jarvis P.Chloroplast biogenesis: Control of plastid development, protein import, division and inheritance.Arabidopsis Book, 2008, 6: e110. |
| [19] | Webber A N, Malkin R.Photosystem I reaction-centre proteins contain leucine zipper motifs. A proposed role in dimer formation.FEBS Lett, 1990, 264: 1-4. |
| [20] | Rutherford A W, Faller P.Photosystem II: evolutionary perspectives.Philos Trans R SocLond B Biol Sci. 2003, 358: 245-253. |
| [21] | Santis-Maciossek G D, Kofer W, Bock A, Schoch S, Maier R M, Wanner G, Rüdiger W, Koop Hans-Ulrich, Herrmann R G. Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology biochemistry and ultrastructure. Plant J, 1999, 18: 477-489. |
| [22] | Peng L W, Yamamoto H, Shikanai T.Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex.Biochim Biophys Acta, 2011, 1807: 945-953. |
| [23] | Andersson I, Backlund A.Structure and function of Rubisco.Plant Physiol Biochem, 2008, 46: 275-291. |
| [24] | Livak K J, Schmittgen T D.Analysis of relative gene expression data using real-time quantitative PCR and the 2 Delta Delta C (T)] method.Methods, 2001, 25: 402-408. |
| [25] | McCouch S R, Kochert G, Yu Z H, Wang Z Y, Khush G S, Coffman W R, Tanksley S D. Molecular mapping of rice chromosome.Theor Appl Genet, 1998, 76: 815-829. |
| [26] | Shi Y F, Chen J, Liu W Q, Huang Q N, Shen B, Leung H, Wu J L.Genetic analysis and gene mapping of a new rolled-leaf mutant in rice (Oryza sativa L.). Sci China C: Life Sci, 2009, 52: 885-890. |
| [27] | Kusumi K, Chono Y, Shimada H, Shimada H, Gotoh E, Tsuyama M, Iba K.Chloroplast biogenesis during the early stage of leaf development in rice.Plant Biotechnol, 2010, 27: 85-90. |
| [28] | Allen J F, Wilson B M, Puthiyaveetil S, Nield J.A structural phylogenetic map for chloroplast photosynthesis.Trends Plant Sci, 2011, 16: 645-655. |
| [29] | Pakrasi H B.Genetic analysis of the form, function of photosystem I and photosystem II.Annu Rev Genet, 1995, 29: 755-776. |
| [30] | 王兴春, 王敏, 季芝娟, 陈钊, 刘文真, 韩渊怀, 杨长登. 水稻糖苷水解酶基因OsBE1在叶绿体发育中的功能. 作物学报, 2014, 40(12): 2090-2097. |
| Wang X C, Wang M, Ji Z J, Chen Z, Liu W Z, Han Y H, Yang C D.Functional characterization of the glycoside hydrolase encoding gene OsBE1 during chloroplast development in Oryza sativa. Acta Agron Sin, 2014, 40(12): 2090-2097. (in Chinese with English abstract) | |
| [31] | Gothandam K M, Kim E S, Cho H, Chung Y Y.OsPPR1, a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis.Plant Mol Biol, 2005, 58(3): 421-433. |
| [32] | Lin D, Jiang Q, Zheng K, Chen S, Zhou H, Gong X, Xu J, Teng S, Dong Y.Mutation of the rice ASL2 gene encoding plastid ribosomal protein L21 causes chloroplast developmental defects and seedling death. Plant Biol (Stuttg), 2015, 17(3): 599-607. |
| [33] | Zhang Z, Tan J, Shi Z, Xie Q, Xing Y, Liu C, Chen Q, Zhu H, Wang J, Zhang J, Zhang G.Albino Leaf1 that encodes the sole octotricopeptide repeat protein is responsible for chloroplast development. Plant Physiol, 2016, 171(2): 1182-1191. |
| [34] | Yoo S C, Cho S H, Sugimoto H, Li J, Kusumi K, Koh H J, Iba K, Paek N C.Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development.Plant Physiol, 2009, 150(1): 388-401. |
| [35] | Wang Y, Zhang J, Shi X, Peng Y, Li P, Lin D, Dong Y, Teng S.Temperature-sensitive albino geneTCD5, encoding a monooxygenase, affects chloroplast development at low temperatures. J Exp Bot, 2016, 67(17): 5187-5202. |
| [36] | Su N, Hu M L, Wu D X, Wu F Q, Fei G L, Lan Y, Chen X L, Shu X L, Zhang X, Guo X P, Cheng Z J, Lei C L, Qi C K, Jiang L, Wang H, Wan J M.Disruption of a rice pentatricopeptide repeat protein causes a seedling- specific albino phenotype and its utilization to enhance seed purity in hybrid rice production.Plant Physiol, 2012, 159(1): 227-238. |
| [37] | Klinghammer M, Tenhaken R.Genome-wide analysis of the UDP-glucose dehydrogenase gene family in Arabidopsis, a key enzyme for matrix polysaccharides in cell walls. J Exp Bot, 2007, 58(13): 3609-3621. |
| [38] | 孙兰茜, 常平安, 曾鑫, 韩丽萍, 冉凤, 王超颖, 王玲, 黄飞飞. 甘油磷酸二酯酶家族蛋白的分子进化. 基因组学与应用生物学, 2015, 34(27): 172-178. |
| Sun L X, Chang P A, Zeng X, Han L P, Ran F, Wang C Y, Wang L, Huang F F.Molecular evolution of the glycerophospho- diesterase family proteins.Genomics Appl Boil, 2015, 34(27): 172-178. (in Chinese with English abstract) |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||